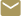
Analyzing the faint electromagnetic signals emitted by biomolecules could give researchers a faster and simpler way to predict how small molecules such as those found in medicines may bind with receptors within cellular membranes. Currently, this binding process is typically predicted using molecular dynamics simulations that require powerful computers and trade accuracy for speed when calculating interactions at greater distances.
Under a Defense Advanced Research Projects Agency (DARPA) program, GTRI researchers have introduced a matched filter technique – similar to what is used to analyze signals returned to radar systems – that uses electrical signals within living cells to predict these molecular binding events. This research has clarified how medications that may be used to treat the disease cystic fibrosis will bind to the membranes that control fluid concentration in the lungs. Beyond cystic fibrosis – a genetic disorder of the lungs and the digestive system – the technique could have broader applications to challenges such as opening pores in biofilms so antibiotic drugs can treat underlying bacterial infections.
Collaborators from the Georgia Tech Research Institute (GTRI), Emory University, the New College of Florida, and the Georgia Tech School of Electrical and Computer Engineering are collaborating on the research. The four-year project, known as Signaling and Electromagnetic ANalysis in the Cellular Environment (SEANCE), is part of DARPA’s larger RadioBio program.
Leveraging Techniques from Radar Signal Processing
“A significant outcome of this program is an analysis technique that leverages signal processing concepts familiar to electrical engineers to describe molecular binding events,” the researchers wrote in a summary of their work. “The team developed the matched filter Molecular Ambiguity Function (MAF), conceived by Georgia Tech School of Electrical and Computer Engineering Professor William Hunt, to quantify the correlation of fields between two molecules. This is analogous to the radar Ambiguity Function, which correlates a returned radar signal to the transmitted signal for detection.”
The MAF provides a measure of binding affinity between two molecules – a ligand and receptor – and relies only on measurement of the local electrical fields, which result from molecular charge, to predict the locations where binding will take place, explained Doug Denison, director of GTRI’s Advanced Concepts Laboratory and the project’s co-principal investigator.
These predictions have traditionally been done using full molecular dynamics simulations, along with evaluation of mechanical interactions between molecules. Molecular dynamics simulations require complex computer algorithms based on an understanding of the basic physics principles governing interactions between the molecules.
In radar signal processing, a matched filter correlates a known original signal with an incoming unknown signal to detect the presence of a distorted version of the original signal. This is useful in radar, for instance, where signals reflected from an aircraft in flight will include a distorted version of the radar waves originally transmitted toward it.
The researchers applied that concept to the biological world, studying the electric fields emitted by molecules of interest. But instead of a radar and reflection from an aircraft, the application is a ligand – a protein molecule – and a receptor to which the ligand binds. The binding typically triggers an activity in the cell containing the receptor.
A New Way to Study Molecular Binding
These ligand-receptor binding events have been studied from both chemical and mechanical standpoints. Mechanically, the binding is similar to a key in a lock where a pocket in the receptor is a perfect fit for the ligand protein. But how do the ligand and receptor find one another?
“That’s really where we zeroed in,” said Denison. “We looked at the electric fields that are just on the surface, just a few Angstroms (An Angstrom is one hundred millionth of a centimeter) beyond the surface of the proteins. And we analyzed those two fields to show that there was a strong correlation for favorable binding orientations and a poor correlation where binding is not going to happen. This has a lot of implications for understanding binding events, and calculating the probability of binding events.”
Based on their new findings, the team was able to study different potential orientations for the ligand-receptor binding to determine the most favorable binding positions between the two molecules. “This has significant and real applications,” Denison said.
Finding Applications in Cystic Fibrosis
Among those potential applications is cystic fibrosis, a disease in which the lungs and digestive system can become overwhelmed by mucus formed in the body. For the past four years, the research team has been working with Nael McCarty, one of the nation’s leading cystic fibrosis researchers and a professor at Emory University.
The research team applied the new MAF technique to a molecule that governs fluid flow across cell membranes. The Cystic Fibrosis Transconductance Regulator (CFTR) functions as a chloride channel, binding to and hydrolyzing an activator molecule – adenosine triphosphate (ATP) – to form a pore that provides a conductive pathway predominately for chloride ions. A dysfunction in the CFTR, induced by mutations, is responsible for cystic fibrosis.
“Using this approach, the team discovered that certain regions of the CFTR molecule surface exhibited anomalously strong interactions with ATP,” the researchers wrote. “These regions are of interest not only because they may possibly have a functional role, but also because they may also serve as communication sites that orient ATP in its binding functions with CFTR.”
Using a series of precise cellular-level electrical signal measurements done in McCarty’s lab, the researchers were able to model the current flows created by the ATP by calculating the extent to which molecular binding caused the channels to open.
Beyond the opening of pores to allow chloride ion flow, the modeling could also help researchers understand how binding events affect another key pathology in cystic fibrosis patients – the development of biofilms that make treating bacterial lung infections more difficult. Improved ability to treat such infections, which are not related to CFTR binding, could benefit people with cystic fibrosis, Denison said.
Working with McCarty, the GTRI researchers plan to continue building on their collaboration around CFTR, with the RadioBio results informing future analysis and experimental studies.
Broader Applications for Designer Molecules
The research may have broader implications for assessing applications for new types of designer molecules being developed to counter disease at the molecular level. Molecular dynamics simulations now help researchers predict how the molecules will bind, but the MAF could provide a new tool to make the overall process faster and more efficient.
“We can rapidly explore a binding space once we know the charge distribution over the molecule and its conformation,” Denison said. “We predict the fields from that. You have charges, a cloud of charge, and that creates a field. Based on that, we can predict the interaction potential.”
The researchers have used graphics processing units (GPUs) to quickly analyze electrical fields to evaluate the binding potential. Having that information might allow researchers to decide when to narrow the focus by applying the more computationally-intensive computer simulations.
“What we are doing can be more efficient than molecular dynamics simulation, which is a really complicated tool,” Denison said. “Molecular dynamics includes a lot of different physical phenomena, from electrostatics and Van der Waals forces (a distance-dependent interaction between atoms and molecules) to thermodynamics and mechanical spring terms. All of these different terms must be calculated on femtosecond (a quadrillionth of a second) time scales to watch the molecules evolve. We now have a way to provide more of an approximate solution that will get us quite far and then allow molecular dynamics to come in at the end – a big hammer to get the full physics.”
Enhancing the Study of Molecular Binding
The techniques developed by the SEANCE research team may be advantageous at interaction ranges of five to ten Angstroms, where full molecular dynamics simulations become more costly. While these distances seem minuscule, they can be beyond the range of certain atomic-scale forces that are important to the molecular binding.
The extent of the “long-range” correlations is reduced by screening effects of water and salts that exist between the molecules of living systems, so the researchers have included the screening effects in their calculations and have found that significant correlations exist before the molecules contact one another.
“When we plotted the correlations in the water outside the proteins, we found energy gradients – forces – directing the ATP molecules toward regions of the surface,” said Ryan Westafer, the project’s other co-principal Investigator. “The underlying physics of the problem provided a common basis for communication with our colleagues spanning multiple disciplines.”
Research in the basic biological sciences is a relatively new direction for the GTRI research team, which has focused on basic electromagnetic science for its traditional work with radar and antenna systems. Denison believes their experience and expertise can provide new and useful perspectives to researchers trained in the biological sciences.
“I think we’ve made some progress in this area by bringing to it a different view from the perspective of electrical engineers,” he said. “By bringing in expertise in electromagnetics, we helped develop a unique capability and toolset that we expect will help our collaborators in the biological sciences.”
In addition to those already mentioned, the research included the following contributors: Kenneth W. Allen, Jonathan Andreasen, Mark Bolding, James Dee, Daniel Dykes, John Farnum, Michael Farrell, Kellie McConnell, Margaret Panetta, Jonathan Perez, Tabitha Rosenbalm, Alex Saad-Falcon, and Nicholas Witten from GTRI; Guiying Cui from Emory University, and Steven Shipman from the New College of Florida.
This material is based upon work supported by the Defense Advanced Research Projects Agency (DARPA) under Contract No. HR001117C0124. Any opinions, findings, conclusions, or recommendations expressed in this material are those of the author(s) and do not necessarily reflect the views of DARPA.
Writer: John Toon (john.toon@gtri.gatech.edu)
GTRI Communications
Georgia Tech Research Institute
Atlanta, Georgia USA
MORE 2022 ANNUAL REPORT STORIES
MORE GTRI NEWS STORIES
The Georgia Tech Research Institute (GTRI) is the nonprofit, applied research division of the Georgia Institute of Technology (Georgia Tech). Founded in 1934 as the Engineering Experiment Station, GTRI has grown to more than 2,800 employees supporting eight laboratories in over 20 locations around the country and performing more than $700 million of problem-solving research annually for government and industry. GTRI's renowned researchers combine science, engineering, economics, policy, and technical expertise to solve complex problems for the U.S. federal government, state, and industry.